
Education and Technical Skills
Currently a Mathematical Modeler at Immunetrics, Inc., I received my B.A. in Mathematics from Goshen College in Indiana and graduated with a Ph.D. in Computational Biology from the University of Kansas. After graduation, I worked for two years as a postdoctoral research associate at Los Alamos National Laboratory. My primary expertise is in modeling dynamical systems, typically for biological or physiological applications.
I am proficient with the Python programming language and am familiar with Java, Scala and C/C++. I regularly use Python scientific libraries (numpy, scipy, pandas, etc.) for modeling and data management. My daily workflow includes numerical integration of systems of ODEs, optimization and validation of models with clinical/experimental data, and analysis and graphing of results.
Research Experience and Interests
My current work involves predictive modeling of mid-to-late phase clinical trials to assess the viability of drug candidates. These disease-specific models often span multiple scales, from intracellular to inter-organ systems. In some cases they merge both mechanistic and statistical modeling approaches.
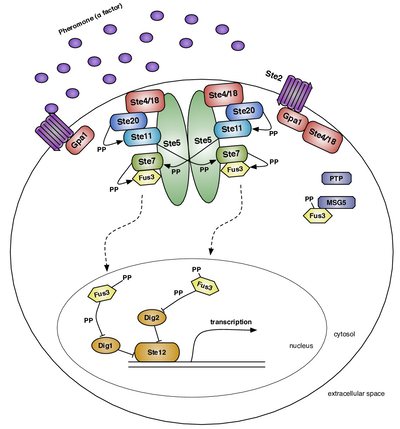
Formerly a member of the Center for Nonlinear Studies and the Theoretical Biology and Biophysics Group at Los Alamos National Lab, I built and analyzed dynamical models of intracellular signaling networks using a variety of methodologies, including rule-based modeling languages (domain-specific languages for modeling biochemical reaction networks) such as Kappa and BioNetGen.
Image from Suderman & Deeds (2013) PLOS Comp Biol 9 (10) e1003278
Tool Development for Scientific Research
Performing computational research often requires custom tools and libraries. I continue to develop and maintain a number of applications designed facilitate data analysis and model construction and optimization for dynamical systems modeling. Projects include:
- EstCC: An application to estimate information theoretic quantities from cellular signaling data
- PyBioNetFit: An application for fitting systems biology models to qualitative and quantitative data
- TRuML: An application for translating between rule-based modeling languages